
In a similar vein, the 1000 genomes project recently published an enormous catalog of variation from almost 1,110 individuals in 14 populations. The Human HapMap project is hard at work on this and recently published a data set with 3.1 million of these SNPs, among 270 individuals from different populations, roughly one every kilobase. It turns out that there are a lot of these single changes.
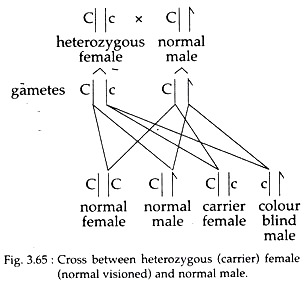
Most studies look at single base changes (SNPs) because they are 1. For an extreme example, look at the Human leukocyte antigen some have hundreds or thousands of alleles, but if you were homozygous for each it would be bad (disease susceptibility) but you'd be fine otherwise.Īgain, it depends on what you consider an allele or locus. Your new/edited question is actually very difficult to answer. Here is a table from this paper which is likewise quite excellent also, check out figure 3. This paper has some AMAZING figures that are worth checking out but are a little more involved. This of course will vary widely from population to population, and amongst outbred individuals.
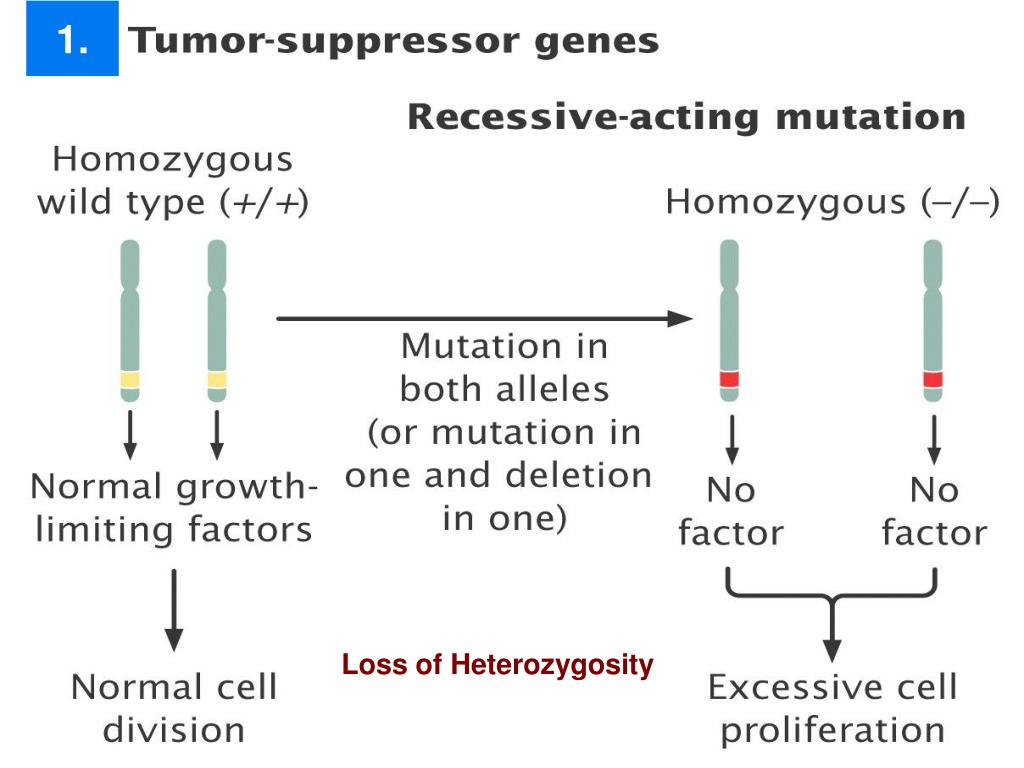
That being said, it turns out there are some serious runs of homozygosity (ROH, meaning long regions of the genome that are homozygous) in human populations. First off, what is your definition of loci? That term is broad, and can be construed to mean everything from large, megabase-length genes to every individual base. If you want an actual number, well, it depends (of course).


 0 kommentar(er)
0 kommentar(er)
